Laboratory of RNA structural functions
RNA typically is a single-stranded biopolymer and is made of ribonucleotides. The self-complementary sequencing within/between RNAs leads to form intramolecular base-pairing structures and intermolecular RNA interactions, respectively. RNA structures and interactions play important roles in many cellular processes, ranging from carrying genetic information, to catalysis, regulation of gene expression, and beyond. Directly determining base pairs or tertiary contacts of RNA structures and interactions are critical steps toward decoding the structural basis of RNA-mediated regulation in cells. To achieve the above purpose, we and others developed a set of novel approaches to determine RNA structures and interactions, based on the principle of crosslinking, proximity ligation, and high-throughput sequencing, such as PARIS2-seq and SHARC-seq. Currently, our lab is interested in combining computational, chemical, and biological approaches to elucidate the fundamental mechanisms of “RNA machines”, including:
Optimize our developed chemical tools to efficiently decode in vivo RNA secondary (2D) and RNA tertiary (3D) structures;
Identify genetic variations that will affect RNA structures;
Investigate the roles of RNA structures and RNA-RNA interactions in human diseases.
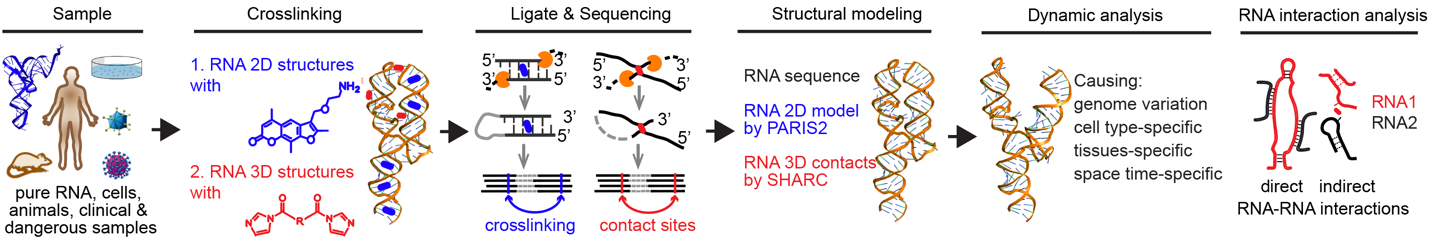
High throughput RNA structure determination methods and their applications.